Different cultivars have different characteristics so correct cultivar identification is important.
An
Australian report relating to olive cultivar identification, ‘Olive Oil
Yield, Quality and Cultivar Identification’ by Kevin Robards and Rod
Mailer, can be found at https://rirdc.infoservices.com.au/items/01-023.
The research described is the start to ongoing work with the aim to
develop DNA testing to identify olive tree cultivars (clones/ near
clones) and to determine relatedness of different olive cultivars (see
dendrogram Fig 5). Cultivars from the Wagga Wagga Olive Grove (WWOG),
situated on the campus of Charles Sturt University and a grove at Yanco
were used in the study. The findings were especially interesting
considering that many trees growing in commercial orchards have been
developed from cuttings taken from WWOG. The study found that some
trees had been incorrectly identified and also that there appears to be
considerable DNA variation within some of the trees previously
identified as like-type cultivars. To fully understand the report we
need to understand how the trees were ‘fingerprinted’. The DNA analysis
procedure used in the study is called RAPD (Random Amplified
Polymorphic DNA).
DNA terms explained (background info only)
Deoxyribonucleic
acid (DNA) is a molecule that contains the instructions for the
development and functioning of living organisms (animals, plants,
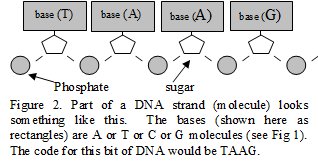
bacteria etc). The DNA molecule is a sugar/ phosphate strand (backbone)
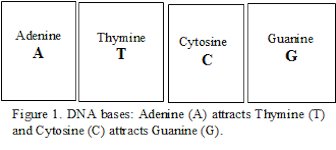
that has atom groups called bases attached (see Figure 1). The order of
the bases is the DNA code (Figure 1 and 2). Adenine (base) attracts
Thymine (base) and Cytosine (base) attracts Guanine. In this way two
DNA strands (the helix) are held together by molecular attraction.
A
gene is a region of DNA that influences a particular characteristic in
an organism. It contains the DNA code used to assemble proteins. Only
about 1.5% of the human genome consists of protein-coding areas.
RAPD DNA testing (fingerprinting)
Related
organisms have similar DNA. DNA testing determines relatedness by
comparing segments of DNA. The RAPD method does not compare all of the
organisms DNA. It does not compare genes. It only compares bits of DNA
that might well not code for anything in particular (protein or
otherwise).
What is compared?
The short segments (pieces or
bits) of DNA that are compared are defined by two segments that
contain specific codes within the DNA. It is not the codes that are
compared but the distance between them and how many times they occur.
The short specific DNA coded sequences define the beginning and the end
of a segment that is compared. The amount (length) of DNA between the
beginning (specific coded region) and end (specific coded region)
varies from organism to organism. The number of times these codes occur
in the DNA also varies organism to organism. The more often these
specific coded areas occur the more segments there will be. Closely
related organisms will have similar numbers of segments of identical
length. Clones (identical organisms) have the same number of identical
length segments.
How do we find these specific codes?
Small
DNA molecules called primers locate the specific codes. Primers are
designed to bind the specific coding regions. The primers act as a
starting point and facilitate ‘*copying’ of DNA material.
How do we find out how many segments there are and how long they are?
To
see the DNA segments with the naked eye you need an awful lot of it. To
make a lot of DNA, the segments are ‘*copied’ many times. This is
called amplification.
* Note: An exact ‘copy’ is not made but a ‘matching copy’. When the ‘copy’ is copied it will be the same as the first.
Equipment required to amplify the DNA segments?
 |
PCR (basic) procedure:
Remove young leaves from the tree.
Separate the DNS from the other leaf cellular molecules.
To the purified DNA add:
primers (short segments of DNA with a specific sequence)
polymerase enzyme (used to copy DNS and
nucleotides (DNA building blocks).
Place mixture in PCR
enter the details (e.g. number of cycles required)
Turn on
|
The
DNA amplification process is called a PCR (Polymerase Chain Reaction)
and it is done in a PCR machine (Figure 3). The PCR machine is an
automated cycler that can quickly heat and cool the reaction mixture.
Each ‘cycle’ doubles the number of DNA segments in the mix. After the
first cycle there are two copies of each segment. After the second
cycle there are four copies of each segment and so on until after 30
cycles you have more than a billion. Each cycle involves three major
steps that are repeated 30-40 times:
Step 1 The mixture is heated so that the DNA helix strands separate.
Step 2 Cooling allows the primer to bind single DNA strands.
Step 3 Warming encourages the enzyme (polymerase) to make DNA using a primer as a starting point.
Separation of segments and measurement
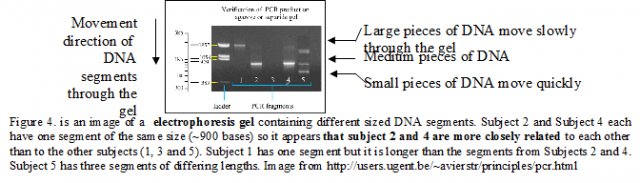
The
amplified DNA is placed in a gel and an electrical charge applied. DNA
contains many phosphorus atoms, which are negatively charged causing
the DNA to be drawn through the gel by the current. Long pieces of DNA
move more slowly than the shorter pieces. The DNA segments are inserted
into the gel together (one lane per organism). As time passes the
shorter strands separate from the longer strands (see Figure 4).
 Setting up a lab
Setting up a labThinking
of setting up a lab in order to test your own trees? Table 1 contains
the prices of some items you will probably require.
Can I have my olive tree DNA tested today?
It
appears that olive DNA fingerprinting is not at this time available in
Australia. Previously olive testing has been done at Wagga Wagga as per
the ‘Olive Oil Yield, Quality and Cultivar Identification’ (OOYQCI)
report (3) and at South Australia’s University of Adelaide’s Waite
Agricultural Research Institute/ South Australian Research and
Development Institute (SARDI). The good news is that Rod Mailer
Principal Research Scientist DPI (Department of Primary Industries)
Adjunct Associate Professor Charles Sturt University at Wagga is
expecting to start another olive tree DNA testing project in the not
too distant future (2007). Rod thinks they will be able to DNA
fingerprint any olive tree for approximately $50 per tree and once they
are “rolling” it should only take a couple of days to process the DNA
and obtain a result (see footnote). As far as I’m aware ‘simple
on-site’ DNA testing of olives and other fruit trees by an average
gardener’ is not available yet. One day (who knows when or how) DNA
field testing using a simple handheld machine will probably be
possible. Don’t hold your breath.
 Summary
SummaryThe
RAPD technique doesn’t provide information regarding the exact
characteristic differences between the trees tested just whether there
are differences. This is because only random areas of selected DNA are
compared not actual genes. Those conducting the tests do need to have
access to the ‘true variety’ ‘markers’ in terms of the lengths and
number of different segments that will be amplified using specific
primers. If you have two unknown plants and want to know if they are
clones, no information is required. RAPD can do the job by comparing
the segments amplified from the two DNA samples (eg. young leaves) you
provide. Any organism (plant, animal etc) that has DNA can be tested
using the RAPD technique.
FootnoteJust before this
article went to print we contacted Rod Mailer. He provided us with Ref
4 and advised us that this year the ‘microsatelite’ technique would be
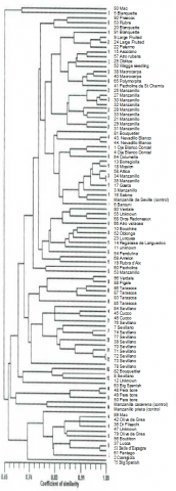
used to fingerprint trees. It should be more useful than RAPD. The
dendrogram (Fig 5 from Reference 4) is a fine example of how important
and useful the research (Ref 3 and 4) has been in terms of olive
cultivar identification and relatedness determination. Trees on the
same branch are similar. You can see for example that all of the
Sevillano trees are on the same branch. The closer the branch is to the
right of the page the more related the two cultivars. Apparen.tly other
publications have not produced a dendrogram as successful as that in
Fig 5.
References
1. http://www.dumru.mc.duke.edu/rapd.html
2. http://users.ugent.be/~avierstr/principles/pcr.html
3. https://rirdc.infoservices.com.au/items/01-023
4.
Mailer, R.J. and May, C.E. 2002. Variability and interrelationships of
olive trees and cultivars using RAPD analysis. Advances in
Horticultural Sciences. (16)3-4: 192-197.





